How to make a shareable code workflow for reproducible and efficient science
Dax Kellie, Martin Westgate



@daxkellie
The replication crisis
Psychology in crisis?

Of 100 replicated studies, only 39 replicated the original result
Biology & ecology in crisis?
Evolutionary biology & ecology research is not immune to wider issues of scientific research
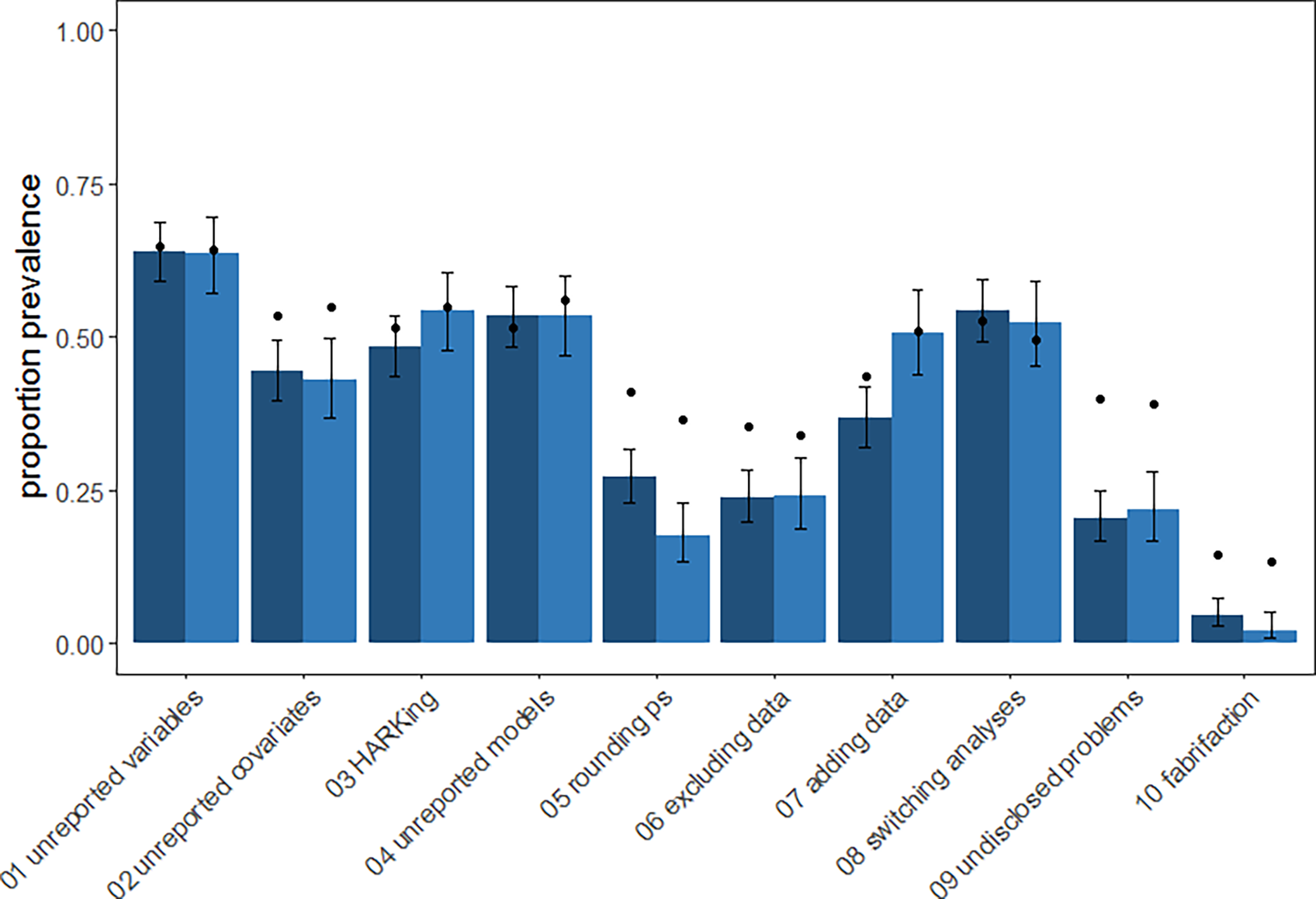
Questionable Research Practices are prevalent but usually unintentional
Reproducibility
Available data and code?

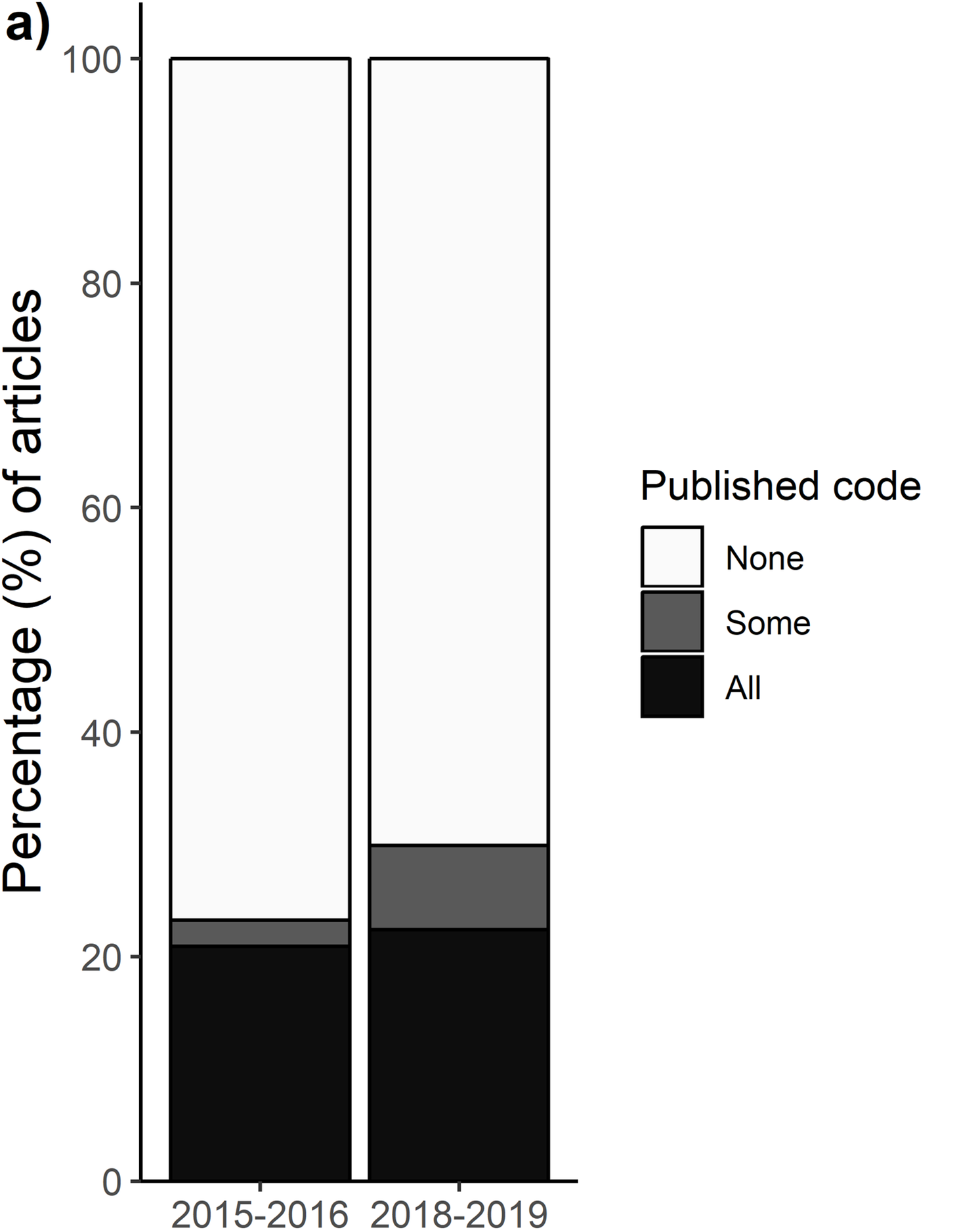
Without knowing how someone cleaned & edited their data, even with data, reproducing a result is difficult
Available code doesn’t guarantee reproducibility
Of 62 Registered Reports from Center of Open Science database:
- 37 had scripts available (60%)
- 31 scripts could be run
- 21 could reproduce main results
(34% of total, 57% of scripts)
Reproducibility is hard

Supporting reproducibility at the ALA
ALA Labs

Supporting reproducibility at the ALA
ALA Labs
The invisible workload of open research
With growing pressure from peers but little change from institutions of how research output is judged, open science is an intimidating amount of work
Small steps
to improve reproducibility*
with big impacts
*of code, mostly
1. Aim to make your work environment shareable
When loading a work environment…
R Projects


R Projects
Without R Project
With R Project
The {here} package
Works nicely with R projects
[1] "C:/Users/DaxKellie/OneDrive/Documents/ALA/Talks/ESA2023"Makes safe file paths easy
[1] "C:/Users/DaxKellie/OneDrive/Documents/ALA/Talks/ESA2023/images/folder/subfolder"The {renv} package
Initialise a new project-local R library
Save the state of the project to a lockfile
{
"R": {
"Version": "4.2.3",
"Repositories": [
{
"Name": "CRAN",
"URL": "https://cloud.r-project.org"
}
]
},
"Packages": {
"markdown": {
"Package": "markdown",
"Version": "1.0",
"Source": "Repository",
"Repository": "CRAN",
"Hash": "4584a57f565dd7987d59dda3a02cfb41"
},
"mime": {
"Package": "mime",
"Version": "0.7",
"Source": "Repository",
"Repository": "CRAN",
"Hash": "908d95ccbfd1dd274073ef07a7c93934"
}
}
}Use GitHub
Backup your project online, share your project with others

Use GitHub
README at the front to explain high-level context, structure, metadata
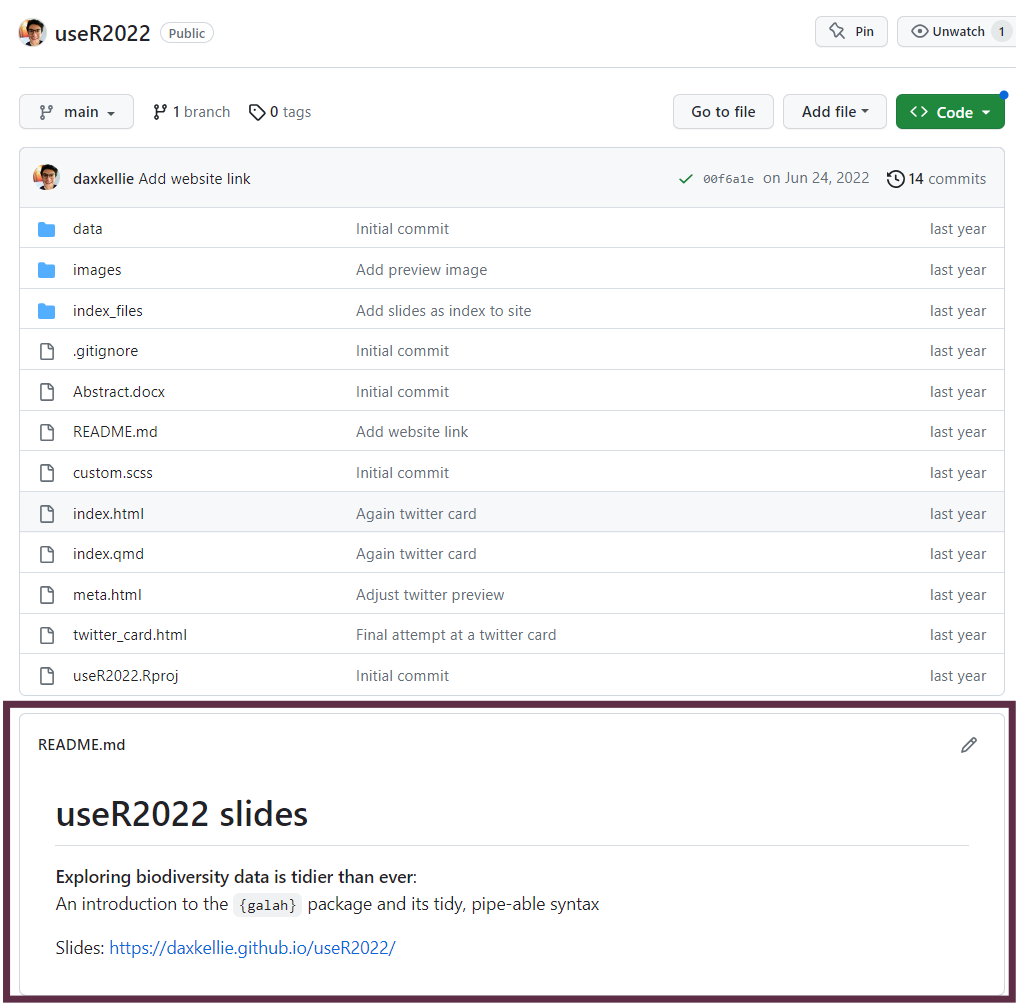
Use GitHub
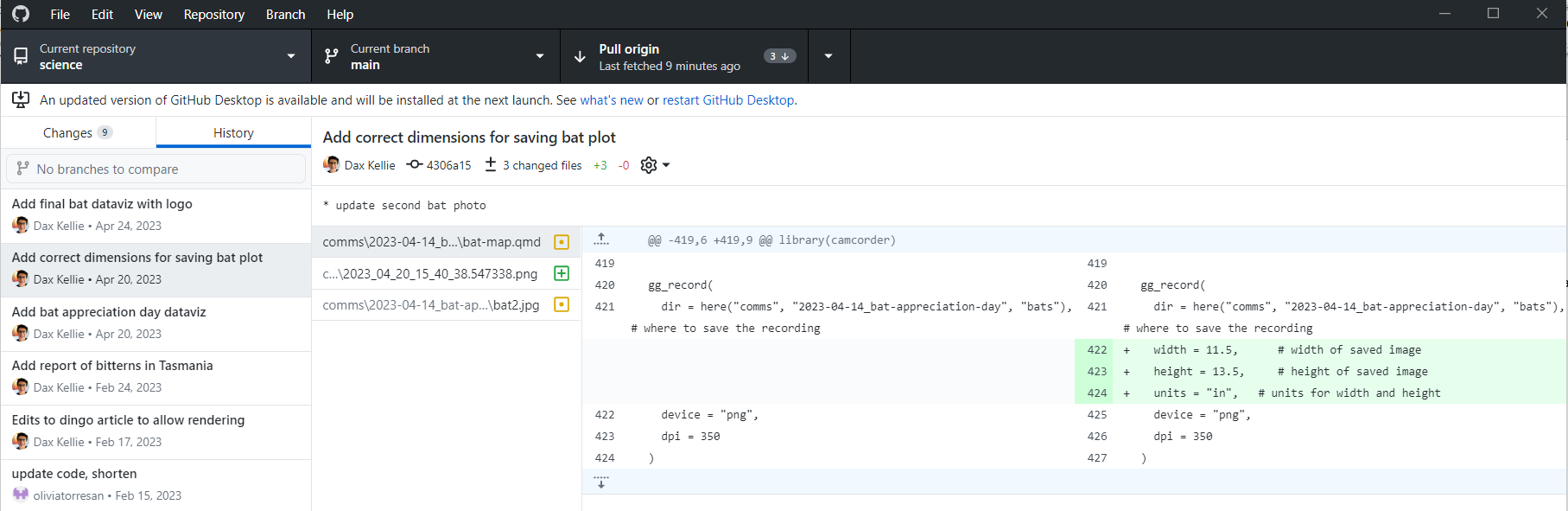
Version control with GitHub Desktop is easy to learn because it’s visual
2. Readable code, readable notes
Simple, clear names
Simple, clear notes (with interpretations)
Meh
Yay
# Test effects of temperature and rainfall on species richness
model <- lmer(outcome ~ predictor_1 + predictor_2 + covariate_1 +
(1|covariate_2) # random effect
data = data)
summary(model)
# Results show significant effect of predictor_1. This suggests [interpretation]...
# However, confidence intervals of significant effect are wide3. Render your code into a document
(with middle steps included)
Document results for easy referencing later
Quickly reference and share your work (because you don’t need to rerun your code)
Document results for easy referencing later
Quarto is like a refined, updated R Markdown - it’s easy and makes documents look nice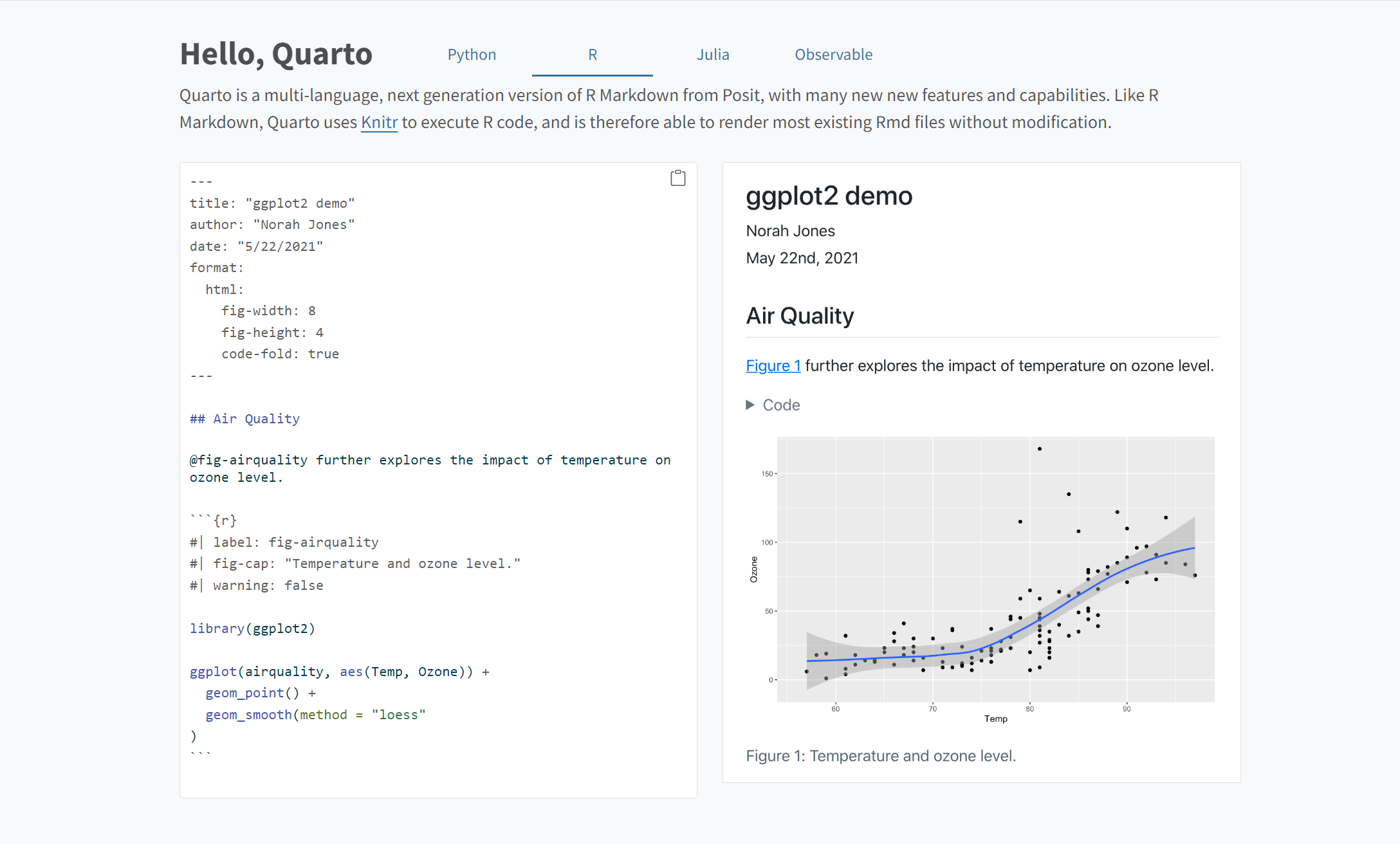
Document results for easy referencing later
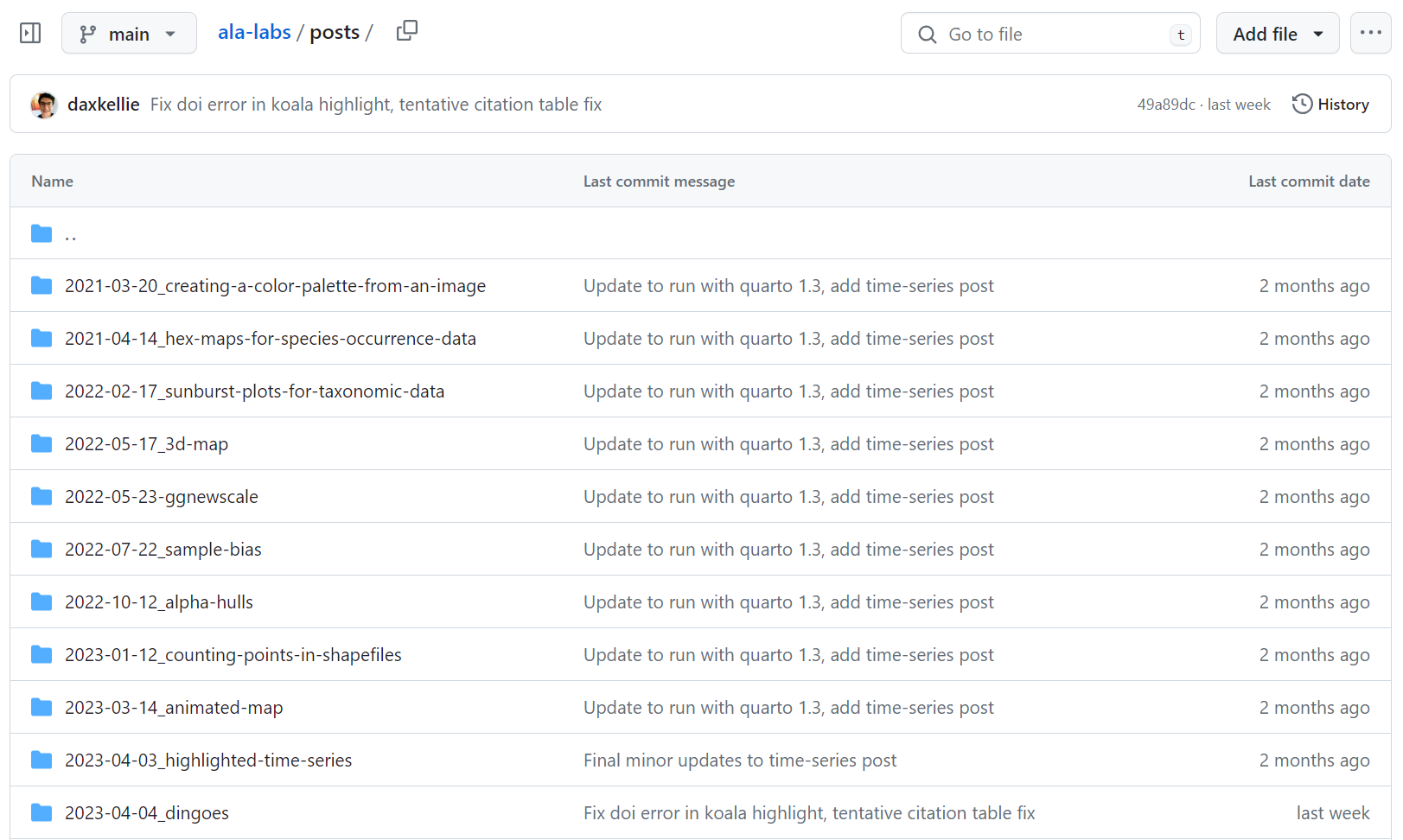
Saving them in one place creates an accessible library of usable code.
This can be public or private
Summary
Small steps to improve reproducibility with big impacts
- Aim to make your work environment shareable
- Create projects with safe links (R Projects + {here})
- Save package versions ({renv})
- Use an online repository (GitHub)- Readable code, readable notes
- Simple, clear object & function names
- Clear notes with interpretations of results- Render your code into a document
- Quickly reference your work (because you don't need to rerun your code)
- Save rendered files somewhere findable/shareable to reference later
- Quarto makes this easier than everThank you
Dax Kellie
Data Analyst & Science Lead
Science & Decision Support | ALA
: dax.kellie@csiro.au
: @daxkellie
: @daxkellie
Science & Decision Support team
Martin Westgate, Fonti Kar, Olivia Torresan
Shandiya Balasubramaniam, Amanda Buyan
Juliet Seers, Callum Waite
These slides were made using Quarto & RStudio
Slides:




